| Top Software Resources
Fun Archives
|
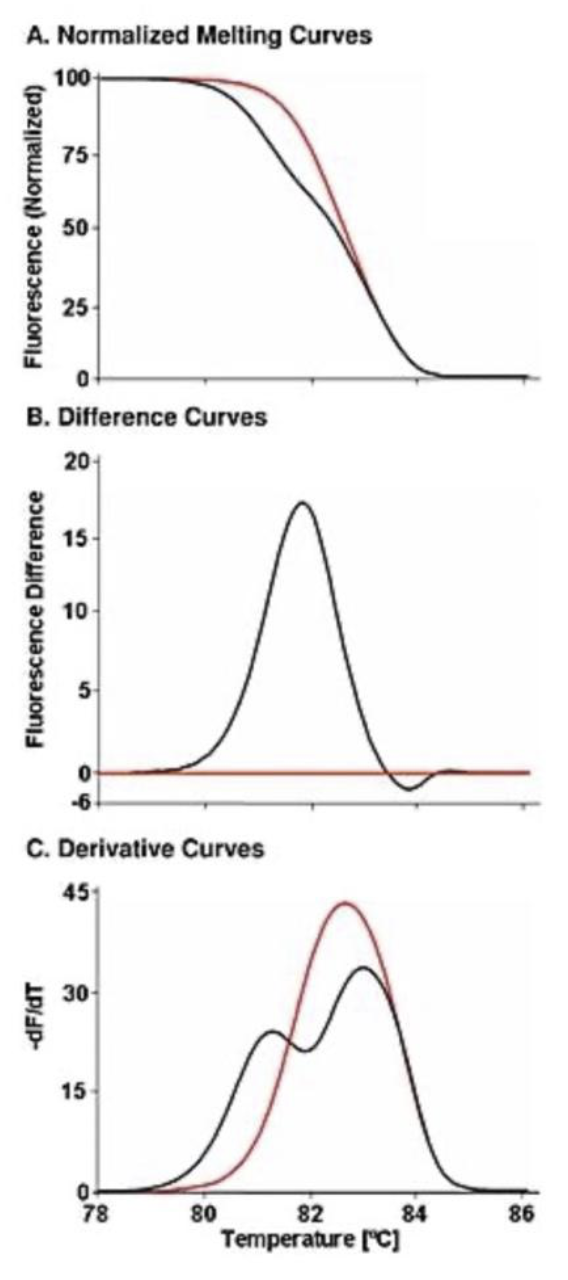
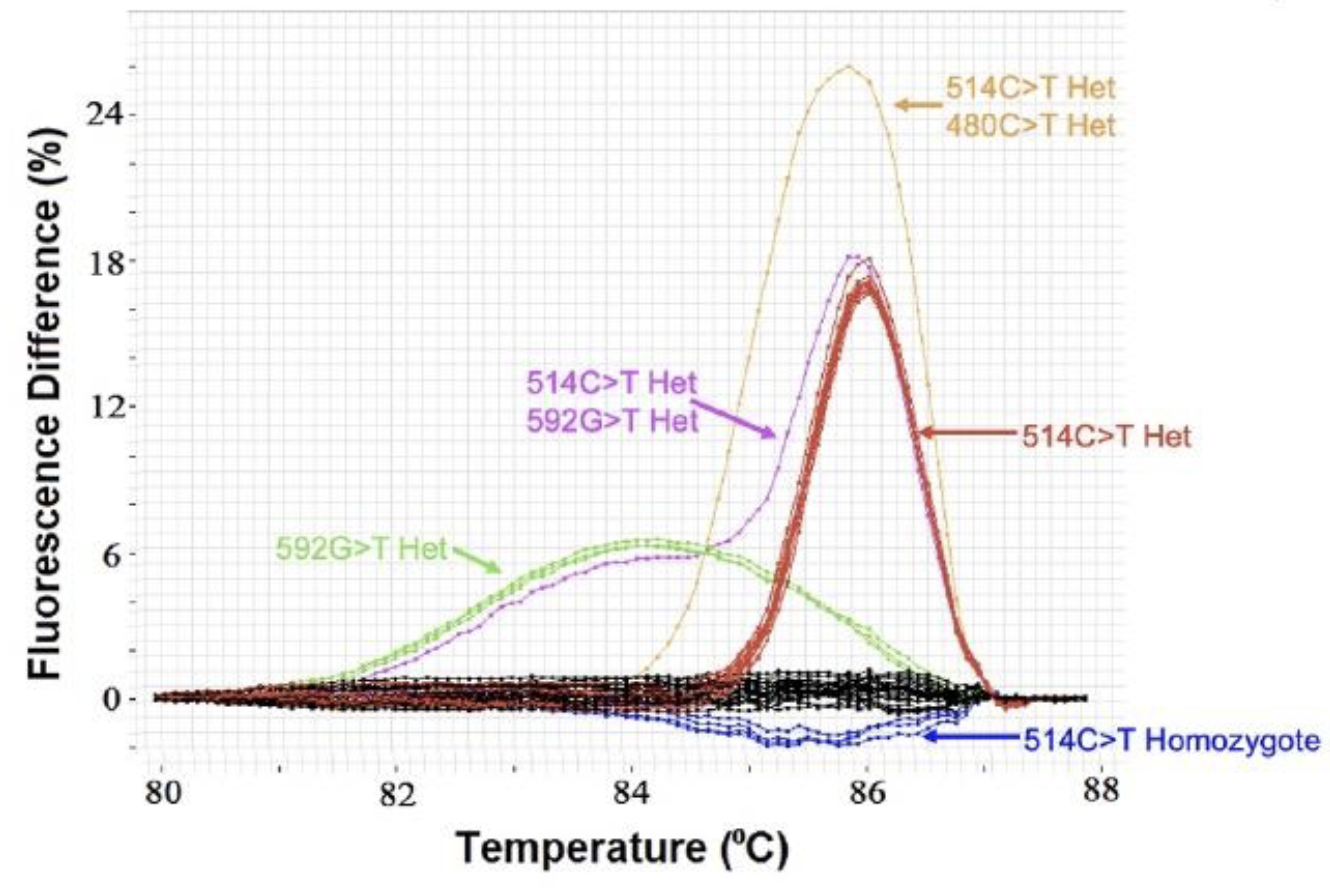
Although identification of DNA variation within an individual is now dominated by massively parallel sequencing (NGS), gene scanning techniques are still useful to screen for variants within a PCR amplicon across individuals. Instead of sequencing all samples, normal samples at limited loci can be identified without sequencing. Legacy techniques such as SSCP, DGGE, dHPLC, and TGCE are seldom used today. Unlike these methods (and NGS), high resolution melting is closed tube and does not require processing, reagent addition, or separation after PCR. High resolution melting detects heterozygotes (mismatched duplexes) that form after amplification of heterozygous DNA and distort the melting curve shape. To accurately compare curve shapes, melting curve data are: 1) normalized to reflect helicity, with adjustments to make fluorescence horizontal before and after the melting transition, 2) overlaid at high temperature to correct for minor temperature offsets, and 3) subtracted from the average of WT curves. These difference plots should not be confused with negative derivative plots. The sensitivity and specificity of heterozygous scanning for SNPs are both 100% for amplicons shorter than 400 bp. For PCR products between 400 and 1,000 bp, the sensitivity is 96.1% and specificity 99.4% (Reed & Wittwer, 2004).
| ||||||||||||||||||||||||||||
| © 2006- Wittwer Lab. All Rights Reserved. |